The Serrapilheira/ICTP-SAIFR Training Program in Quantitative Biology and Ecology will train young Brazilian and other Latin American scientists for world-class research on biology and ecology using the quantitative methods of mathematics, physics, and computer science. It strategically makes use of the country’s well-established excellence in mathematics and physics to tap into Brazil’s tremendous potential in life sciences.
Brazil’s ecosystems are among the richest in the world, and it is becoming increasingly clear that their inherent complexity poses enormous scientific challenges to find solutions that allow them to survive and flourish. The goal is to build an interconnected network of highly skilled scientists who can make significant contributions to advance the field in Brazil and Latin America.
To reach its goal, the training program will be highly selective and involve students at the beginning of their graduate studies who have already developed quantitative skills and are interested in applying these skills in solving cutting-edge problems in biology and ecology. Lectures in all areas of biology and ecology will be presented by international experts, and no previous knowledge of biology is required.
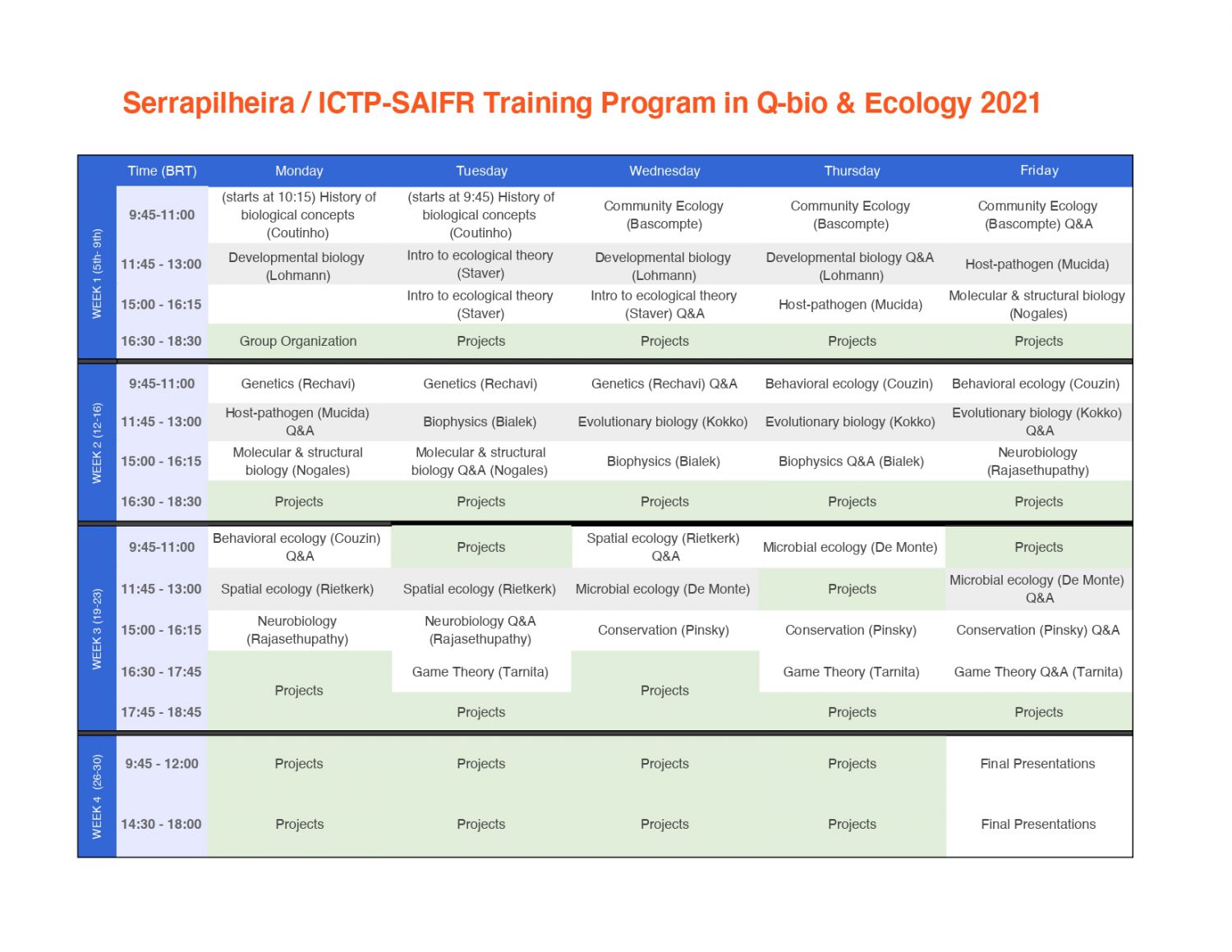
Because of the COVID-19 pandemic, the first edition of the program will take place in a shorter online version during the period of July 5-31, 2021. There were online keynote presentations via Youtube on March 2 by Prof. Simon Levin (Princeton U.) (click here to watch) and on March 4 by Prof. Akiko Iwasaki (Yale U.) (click here to watch) to announce the program. Future editions of the training program will take place at ICTP-SAIFR/IFT-UNESP in São Paulo with the second edition planned for the 6 month period of July – December, 2022.
Students with quantitative skills who will have finished their undergraduate studies by the end of 2021 at a Latin-American institution can apply to participate in the four-week online training program by reading the application instructions and submitting the online application form before May 3, 2021 at 17:00 BRT.. Approximately 50 students will be accepted into the program, and priority will be given to the first 500 applications received. Confirmed lecturers in the July program are listed on this webpage, and any questions should be sent to qbioprogram@ictp-saifr.org
In addition to attending lectures by internationally renowned researchers, students in the program will work in small
groups on a research project supervised by a postdoctoral mentor. Applications are open until April 8, 2021 for postdoctoral mentors who will be responsible for proposing and supervising a one-month research project. Mentors should have a PhD in an area related to quantitative biology and can apply using the online form on this webpage. The mentor’s responsibilities will include participation in the first stages of the student selection process, acting as chair in some lectures, and supervising and mentoring the research project for a small working group of 4-5 students. Accepted mentors will receive a single 4000 BRL stipend and are invited to participate in all online lectures during the program.
1. Antonio Coutinho (Instituto Gulbenkian de Ciência) – History of biological concepts
Antonio Coutinho is an immunologist with an extensive career and a comprehensive view of science and scientific thinking. In addition to leading groups and institutions in Sweden, Switzerland, and France, from 1998 to 2012 he directed the Instituto Gulbenkian de Ciência, in Portugal, considered one of the best research training centers in the world.
2. Oded Rechavi (Tel Aviv U.) – Genetics, epigenetics and large datasets
Oded Rechavi works on the transgenerational inheritance through epigenetic mechanism involving small RNAs.
Recent publications:
Three Rules Explain Transgenerational Small RNA Inheritance in C. elegans. Houri-Zeevi L et al. Cell. 2020.
Neuronal Small RNAs Control Behavior Transgenerationally. Posner R et al. Cell. 2019.
A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Houri-Ze’evi L et al. Cell. 2016.
3. Hanna Kokko (U. of Zurich) – Evolutionary biology
Hanna Kokko works on evolutionary ecology of sexual and asexual reproduction, analysis and management of animal populations, evolution of reproductive and social strategies, and sustainability science.
Recent publications:
Optimal germination timing in unpredictable environments: the importance of dormancy for both among- and within-season variation. Ten Brink H et al. Ecol Lett. 2020.
Transmissible cancers and the evolution of sex under the Red Queen hypothesis. Aubier TG et al. PLoS Biology 2020.
The rate of facultative sex governs the number of expected mating types in isogamous species. Constable GWA et al. Nat Ecol Evol. 2018.
4. Eva Nogales (HHMI/U. California at Berkeley) – Molecular, structural and cell biology
Eva Nogales studies macromolecular assemblies of whole units of molecular function by direct visualization of their architecture, functional states, and regulatory interactions using state-of-the-art cryo-electron microscopy (cryo-EM) and image analysis, as well as biochemical and biophysical assays.
Recent publications:
JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications. Kasinath V, et al. Science 2021.
Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Patel AB et al. Science. 2018.
Near-atomic model of microtubule-tau interactions. Kellogg EH et al. Science. 2018.
5. Ingrid Lohmann (U. of Heidelberg) – Developmental biology
Ingrid Lohmann is a developmental biologist, and her team works on the fundamental role of Hox proteins in the process of development of the fruit fly. More specifically, their interest goes from the control of stem cell proliferation to neurogenesis and metabolism during the process of development.
Recent publications:
ATF4-Induced Warburg Metabolism Drives Over-Proliferation in Drosophila. Sorge S et al. Cell Rep. 2020.
Multi-level and lineage-specific interactomes of the Hox transcription factor Ubx contribute to its functional specificity. Carnesecchi J et al. Nature Commun. 2020.
The Hox transcription factor Ubx stabilizes lineage commitment by suppressing cellular plasticity in Drosophila. Domsch K. et al. Elife 2019.
6. Priyamvada Rajasethupathy (The Rockefeller U.) – Neurobiology
Priya Rajasethupathy’s team bridges systems genetics and systems neuroscience to provide unique cross-disciplinary insights into memory. She aims to reveal the molecular, structural, and functional changes governing the evolution of a memory, and ultimately further understand cognitive processes during health and disease.
Recent publications:
A Thalamic Orphan Receptor Drives Variability in Short-Term Memory. Hsiao K et al. Cell. 2020.
Targeting Neural Circuits. Rajasethupathy P et al. Cell. 2016.
Projections from neocortex mediate top-down control of memory retrieval. Rajasethupathy P et al. Nature. 2015.
7. Daniel Mucida (Rockefeller U.) – Host-pathogen interactions and disease ecology and epidemiology
Daniel Mucida studies how the immune system associated with intestinal mucosae maintains a careful balance by generating efficient protective responses without jeopardizing its tolerance to innocuous foreign substances. He will be teaching together with his research team members Angelina M. Bilate and Bernardo Reis.
Recent publications:
Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose. Muller PA et al. Science. 2020.
Adrenergic Signaling in Muscularis Macrophages Limits Infection-Induced Neuronal Loss. Matheis F et al. Cell. 2020.
Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Reis BS et al. Nature immunology 2013.
T Cell Receptor Is Required for Differentiation, but Not Maintenance, of Intestinal CD4+ Intraepithelial Lymphocyte. Bilate AM, et al. Immunity 2020.
8. William Bialek (Princeton U.) – Biophysics
William Bialek works on the dynamics of individual biological molecules, the decisions made by single cells in a developing embryo, and the system that the brain uses in representing information.
Recent publications:
Coarse Graining, Fixed Points, and Scaling in a Large Population of Neurons. Meshulam L et al. Phys Rev Lett. 2019.
Collective Behavior of Place and Non-place Neurons in the Hippocampal Network. Meshulam L et al. Neuron. 2017.
Thermodynamics and signatures of criticality in a network of neurons. Tkačik G et al. Proc Natl Acad Sci U S A. 2015.
9. Silvia De Monte (ENS Paris/Max Planck Inst. for Evolutionary Biology) – Microbial ecology
By combining mathematical models, experiments in controlled conditions and environmental data analysis, Silvia De Monte and her team explore the interplay of cellular-level properties and collective function on the ecological and evolutionary time scales.
Recent publications:
Ubiquitous abundance distribution of non-dominant plankton across the global ocean. Ser-Giacomi E et al. Nat Ecol Evol. 2018.
The evolution of adhesiveness as a social adaptation. Garcia T et al. Elife. 2015.
Can we detect oceanic biodiversity hotspots from space? De Monte S et al. ISME J. 2013.
10. Carla Staver (Yale U.) – Ecology & Introduction to ecological theory
Her work focuses on the dynamics and distribution of biomes, especially within and at the intersection of savanna and forest. Her team uses a combination of empirical and modeling approaches to understand how local interactions of trees with their resource and disturbance environment scale up to predict landscape- and continental-scale patterns in tree cover and the distributions of biomes.
Recent publications:
Spatial patterning among savanna trees in high-resolution, spatially extensive data. Staver AC et al. Proc Natl Acad Sci U S A. 2019.
Forest extent and deforestation in tropical Africa since 1900. Aleman JC et al. Nat Ecol Evol. 2018.
Top-down determinants of niche structure and adaptation among African Acacias. Staver AC et al. Ecol Lett. 2012.
11. Corina E. Tarnita (Princeton U.) – Game theory in ecology and evolution.
Corina Tarnita’s research examines the organization and emergent properties of complex adaptive systems at multiple scales, from single cells to entire ecosystems. Simultaneously, her team uses empirical data to identify and catalog patterns in nature and, within the general frameworks, they develop models whose predictions they attempt to empirically test using eco-evolutionary experiments, molecular and genomic analyses, and field manipulations.
Recent publications:
Eco-evolutionary significance of ‘loners’. Rossine FW et al. PLoS Biology 2020.
Emergence of diverse life cycles and life histories at the origin of multicellularity. Staps M et al. Nat Ecol Evol. 2019.
A theoretical foundation for multi-scale regular vegetation patterns. Tarnita CE et al. Nature. 2017.
12. Jordi Bascompte (U. Zurich) – Community ecology and biodiversity, and ecological networks
Jordi Bascompte combines mathematical models, simulations, and data set analyses to address fundamental and applied questions in ecology. His current major research interest focuses on the structure and dynamics of ecological networks. Jordi is also a member of the Advisory Committee for the Training Program in Quantitative Biology and Ecology.
Recent publications:
Indigenous knowledge networks in the face of global change. Cámara-Leret R et al. Proc Natl Acad Sci U S A. 2019.
Ecological networks. On the structural stability of mutualistic systems. Rohr RP et al. Science. 2014.
The sudden collapse of pollinator communities. Lever JJ et al. Ecol Lett. 2014.
13. Iain Couzin (Max Planck Inst. of Animal Behavior) – Behavioral ecology
Iain Couzin focuses on revealing the principles that underlie collective animal behavior. By developing an integrated experimental and theoretical program, his research aims to understand how, and why, social behavior has evolved in a large variety of systems, from swarming locust, to schooling fish, to flocking birds.
Recent publications:
Individual and collective encoding of risk in animal groups. Sosna MMG et al. Proc Natl Acad Sci U S A. 2019.
Heterogeneous Preference and Local Nonlinearity in Consensus Decision Making. Hartnett AT et al. Phys Rev Lett. 2016.
Revealing the hidden networks of interaction in mobile animal groups allows prediction of complex behavioral contagion. Rosenthal SB et al. Proc Natl Acad Sci U S A. 2015.
14. Max Rietkerk (Utrecht U.) – Spatial ecology
Max Rietkerk’s team has discovered that spatial vegetation patterns in dry ecosystems follow certain mathematical laws, which can provide insight into how close the ecosystem is to a threshold value for sudden desertification. The team studies the mechanisms leading to these patterns, through which they understand how sudden desertification can be prevented and how areas already affected can be restored.
Recent publications:
The effect of climate change on the resilience of ecosystems with adaptive spatial pattern formation. Bastiaansen R et al. Ecol Lett 2020.
Multistability of model and real dryland ecosystems through spatial self-organization. Bastiaansen R et al. Proc Natl Acad Sci U.S.A. 2018.
Self-organized patchiness and catastrophic shifts in ecosystems.. Rietkerk M et al. Science. 2004.
15. Malin Pinsky (Rutgers U.) – Climate change impacts of biodiversity + Conservation, management and decision-making
Malin Pinsky studies population and community dynamics in primarily coastal marine ecosystems with the goal of understanding the impacts of global change and the actions that could foster abundant wildlife and healthy ecosystems. His team uses statistical tools, field ecology, population genomics, and mathematical modeling to understand general patterns that extend across larger spatial scales, deeper in time, and across a wider range of species than would be possible with more traditional techniques.
Recent publications:
Climate-driven shifts in marine species ranges: scaling from organisms to communities. Pinsky ML et al. Annual Review of Marine Science. 2020.
Greater vulnerability to warming of marine versus terrestrial ectotherms. Pinsky ML et al. Nature. 2019.
Preparing ocean governance for species on the move. Pinsky ML et al. Science. 2018.
Youtube link to webinar: https://youtu.be/STQpQx2Q_hg
Youtube link to webinar: https://youtu.be/QMCxo1cXaaE
Click here for Application Instructions in English.
Click here for Application Instructions in Portuguese.
Click here for Application Instructions for Postdoctoral Mentors in English.
Click here for Application Instructions for Postdoctoral Mentors in Portuguese.
Click here for Project Template for Postdoctoral Mentors
Click here for Frequently Asked Questions in English and Portuguese.
Please send any other questions to qbioprogram@ictp-saifr.org
Organizers
Ricardo Martinez-Garcia (ICTP-SAIFR)
Postdoctoral Mentors
Students
Anna Rebello Landim, Rio de Janeiro
Bruna Fistarol, Rio de Janeiro
Bruno Paranhos, Rio de Janeiro
Carlos Andrés Díaz Rodríguez, Colômbia
Daniel Cardoso Pereira Jorge, São Paulo
Denise Cammarota, Argentina
Ewaldo Leitão, USA
Fernanda Thimoteo Azevedo Jorge, Rio de Janeiro
Hemanoel Passarelli Araujo, Minas Gerais
Inês Motta Comarella, Espírito Santo
Irina Lerner, São Paulo
João Pedro Valeriano Miranda, Distrito Federal
José Andrés Guzmán Morán, São Paulo
Luisa Ramirez, Minas Gerais
Maria Julia Maristany, United Kingdom
Matheus Bongestab, Paraíba
Natália Diesel, Portugal
Nathan de Oliveira Silvano, Rio de Janeiro
Noemi Zeraick Monteiro, Minas Gerais
Paula Favoretti Vital do Prado, São Paulo
Pedro Goes Nogueira de Sá, São Paulo
Rafael Menezes, Bahia
Renata Biaggi Biazzi, São Paulo
Tan Tjui Yeuw, São Paulo
Tatiana Campos Neves, Rio de Janeiro
Thiago Moura Rocha, São Paulo
Victor Klein de Sousa, São Paulo